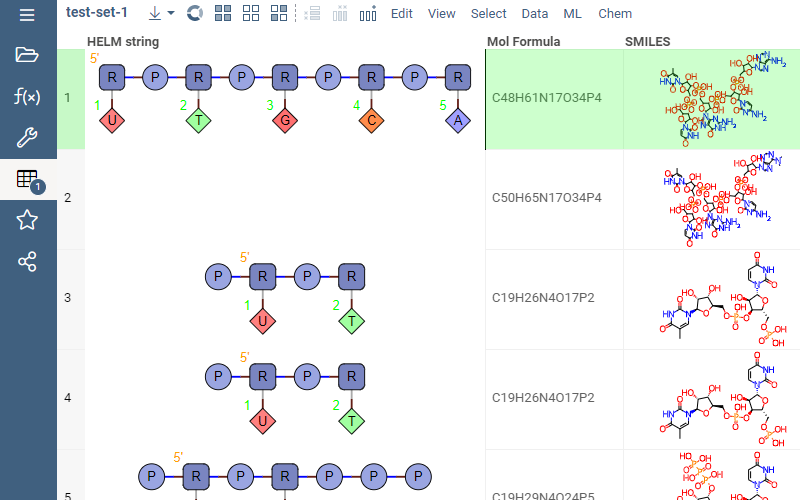
Provides support for HELM notation (Hierarchical Editing Language for Macromolecules) - a single notation that can encode the structure of complex biomolecules including diverse polymers, non-natural monomers and complex attachment points.
Andrew Skalkin | issue tracker | beta
- Automatic detection of HELM values in datasets
- Rendering in the spreadsheet
- Editing
- Conversion to SMARTS and MOLBLOCK
- Substructure search
- Similarity search
- Calculators (mol weight, mol formula, extinction coefficient, )
- Data augmentation (show relevant information in the context panel)
- Integration with the HELM2WebService (calculations, images, etc)
- Support for company-specific monomer libraries
- Management tool for monomer libraries (integrated with the security and privileges)
- Similarity and diversity analyses
- HELM space (UMAP or t-SNE based on the distance)
- Activity cliffs analysis (based on the HELM space)
Kudos to Pistoia alliance for open-sourcing the HELM Web Editor that is used in this integration.
See also: